We’re not all equal in the face of the coronavirus
People across the globe do not all have identical immunity for protecting themselves against the coronavirus. A UNIGE study is comparing and identifying the genetic variants that are potentially the most effective.
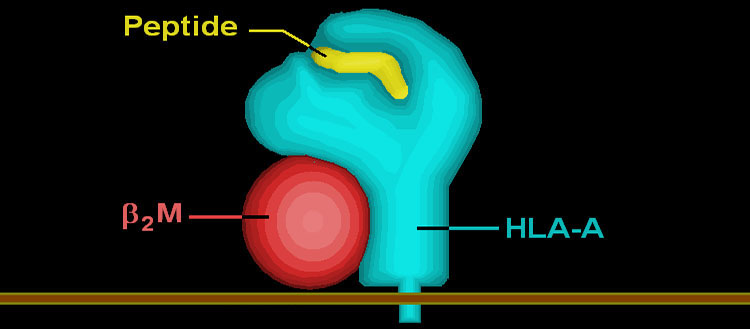
Diagram of a HLA molecule (in blue) with its associated light chain, b2-microglobulin (in red) fixed in the membrane of a human cell and bound to a peptide derived from a virus (in yellow). @DR
Are there differences in immunity to the SARS-CoV-2 coronavirus between populations from different geographic regions? Part of the answer to this question is to be found in the genomes of these groups of people and, more specifically, in the HLA genes responsible for the adaptive immune system. These genes are special in that they often differ between individuals. Thousands of possible variants (or alleles) have been identified, and not all of them are equally effective in fighting a new virus. The frequency of these alleles varies from one population to another due to past migrations and their adaptation to different environments. In a study to be published in the journal HLA, scientists from the University of Geneva (UNIGE) – working in collaboration with the Max Planck Institute in Jena (Germany) and the University of Adelaide (Australia) – have pinpointed the HLA variants that are potentially the most effective against seven viruses, including the new coronavirus. They have also brought to light significant differences between populations.
The genetic variability of immunity lies particularly in the genes of the HLA (Human Leukocyte Antigen) system. These genes produce HLA molecules that are positioned on the surface of cells. When a virus infects an organism, the invader’s proteins are first cut into small fragments called peptides. The HLA molecules then bind on to these fragments and expose them to the surface of the cells, thereby triggering a cascade of immunity reactions designed to eliminate the virus.
Alicia Sanchez-Mazas, a professor at the Anthropology Unit in UNIGE’s Faculty of Sciences, explains: “From the 450 or so most common HLA molecules in hundreds of populations worldwide, we tried to identify the ones that are most strongly bound to the peptides of the new coronavirus.” Over 7,000 peptides can be derived from all of the viral proteins of the coronavirus.
The Geneva-based researcher and her international team used bioinformatic tools to perform the analysis. These can predict the binding affinities between the HLA molecules and the viral peptides on the basis of their physical and chemical properties. The scientists then turned to statistical models to compare the frequencies of these HLA variants in different human populations.
Classification of HLA molecules
The study classified the approximately 450 HLA molecules according to their relative capacity to bind the coronavirus peptides. It provides an essential reference inventory for identifying the genetic resistance or susceptibility of individuals to the virus. The study has also shown that the frequencies of these HLA variants differ significantly from one population to the next.
José Manuel Nunes, a researcher at the Anthropology Unit – and co-author of the article – further explains: “We were surprised to find that Indigenous populations in America had both the highest frequencies of HLA variants that bind the most strongly to the peptides and the lowest frequencies of those that bind the least strongly.” However, as José Manuel Nunes continues, we should not draw too hasty a conclusion from these results: “HLA molecules contribute to the immune response but they are far from being the only element that can be used to predict effective or ineffective resistance to a virus. This is also verified on the ground since America’s Indigenous populations are apparently no less affected than others by COVID-19.”
“Generalist” molecules
In the same study, the authors also analysed the HLA-peptide bindings for all of the proteins of the six other viruses with pandemic potential (two other coronaviruses, three influenza viruses and the HIV-1 virus of AIDS). This showed that many HLA variants are capable of binding strongly to the peptides of all seven viruses studied. Others do the same for all respiratory-type viruses (coronavirus and influenza). This means that there are numerous “generalist” HLA molecules that are effective against a number of different viruses.
“The differences between populations observed in this study are actually differences in the frequencies of the generalist HLA variants that do not bind specifically to the coronavirus but also to other pathogens”, points out professor Sanchez-Mazas. “This is what makes us think that the current differences between populations are the result of past adaptations to different pathogenic pressures, which is extremely informative for understanding the genetic evolution of our species.”
A logical follow-up to the study will be to determine precisely which coronavirus peptides are most strongly bound to the HLA molecules. It is these peptides that will have the highest chances of triggering an effective immune reaction. Identifying them will be vital for developing a vaccine.
