Recherche (en anglais)
ENCODED LIBRARY TECHNOLOGIES
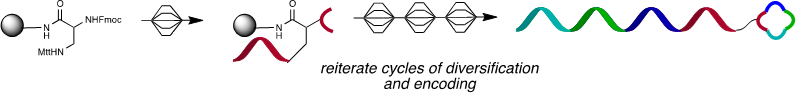
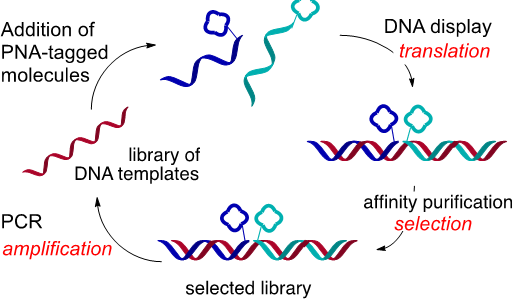
In the field of DNA-encoded libraries, we've pursued peptide nucleic acid (PNA) tagging as a means to encode small molecules and program their assembly based on hybridization instructions (Curr. Opin. Chem. Biol. 2015, 26, 8-15). This method not only offers a simple solution that overcomes the limitation of split and pool combinatorial synthesis techniques but also enables radically new means of screening by selection for the fittest ligand. This technology was used to prepare the largest glycan array reported to date and the first covalent inhibitor of bromodomains that can be used for proteomic profiling. Subsequently, we demonstrated that PNA-tagged molecules could be combinatorially displayed onto DNA libraries, providing a rapid entry into a large molecular diversity space and enabling reiterative cycles of “translation” of DNA into a library of drug-like small molecules, selection and amplification by PCR. These technological developments enabled the discovery of an assembly that can outcompete the interaction of HIV with dendritic cells and the most potent inhibitor of HSP70 reported to date.
PNA-encoded synthesis (PES) using split and mix technique
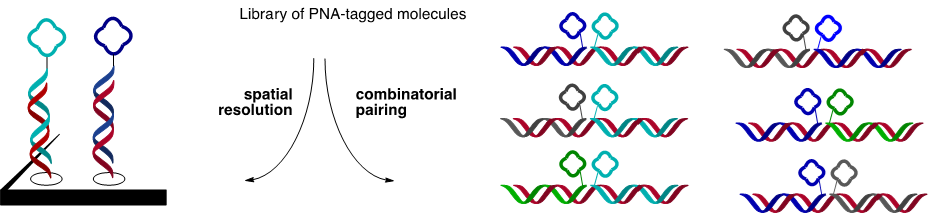
DNA Display:
- Self assembly of PNA-tagged ligand (blue) onto DNA template (red) with control over interligand distance and oligomer order
 - Mixtures of PNA-tagged molecules can be spatially resolved on microarrays and combinatorially paired onto library of DNA templates
- Mixtures of PNA-tagged molecules can be spatially resolved on microarrays and combinatorially paired onto library of DNA templates
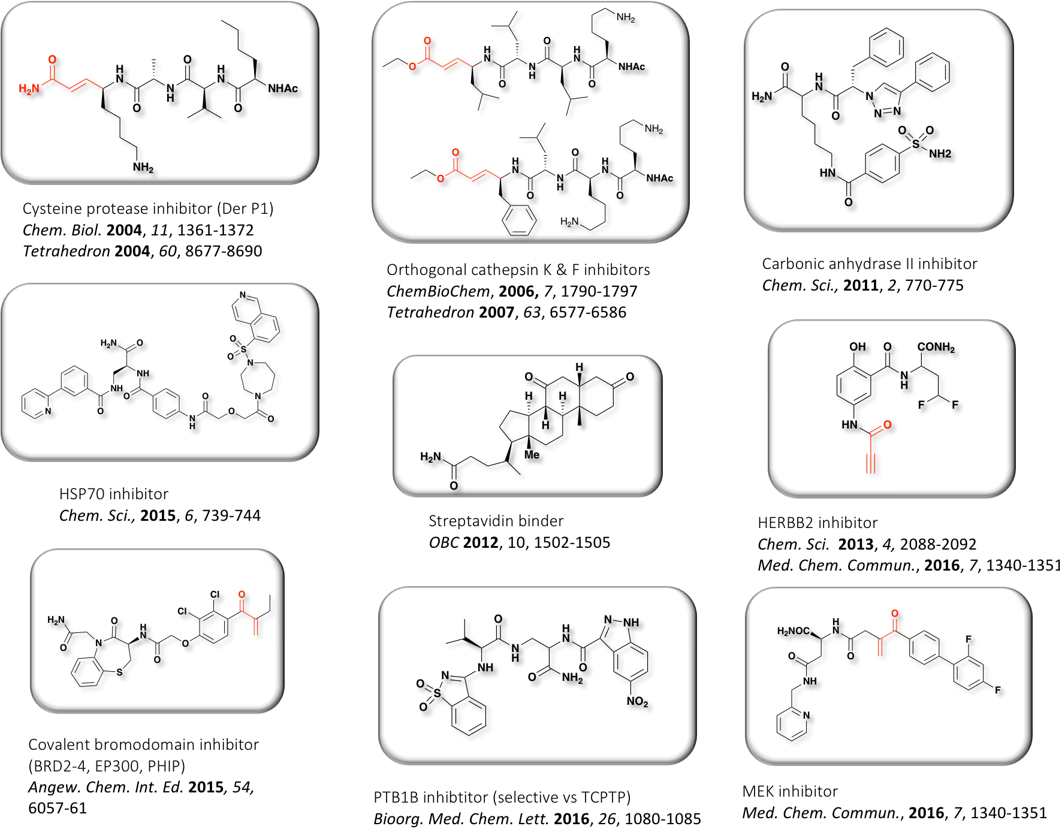
Representative examples of hits identified with PNA-ended libraries
Schematic representation of iterative cycles of selection, amplification and translation
REACTIONS TEMPLATED BY BIOSUPRAMOLECULAR INTERACTION
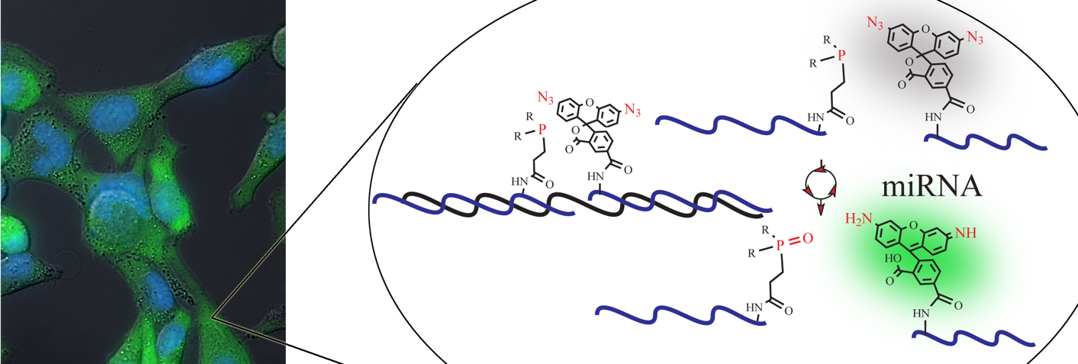
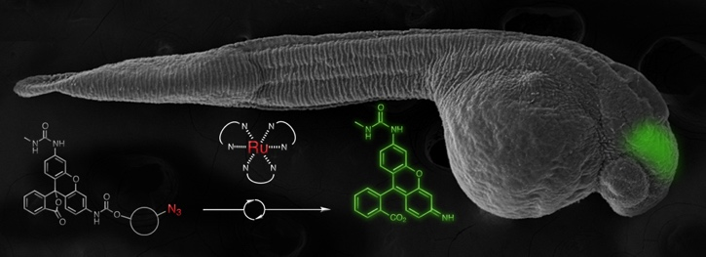
We have also developed several new DNA or RNA templated chemical reactions that proceed in live cells and can be used to image cellular nucleic acid or trigger the release of a bioactive small molecule. This work led to the development of the first azide-based profluorophores and highlighted the power of ruthenium complexes [Ru(bpy)3 analogs] to carry out a photocatalytic bioorthogonal transformations in cellulo and in live vertebrates. We further coupled the photoexcitation of ruthenium complexes to bioluminescence to engineer abiotic transformations. Throughout this work, we have pioneered the use of PNA as a biosupramolecular tag to program assemblies in chemical biology (Acc. Chem. Res. 2015, 48, 1319-1331).
Imaging of cellular miRNA using templated Staudinger reaction
Imaging of nucleic acid in zebrafish
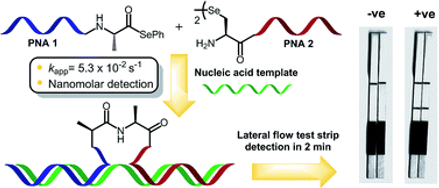
Detection of miRNA with a lateral flow assay
Detection of dsRNA-duplexes using a tempaled reaction with probes forming triplexes
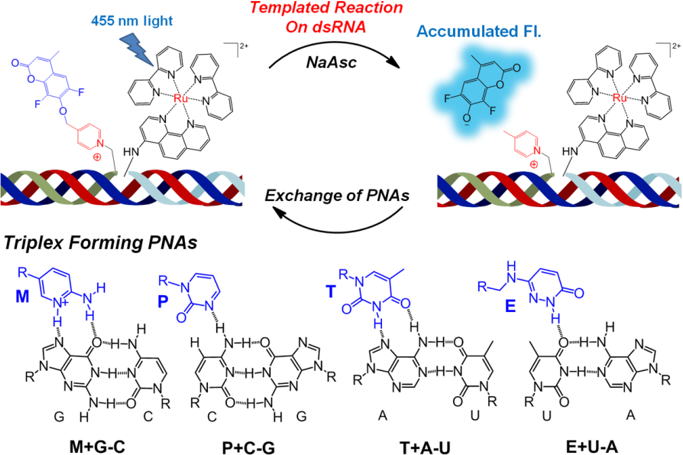
Using a molecular beacon to mask a catalyst in fast templated reaction leverage on short sequences
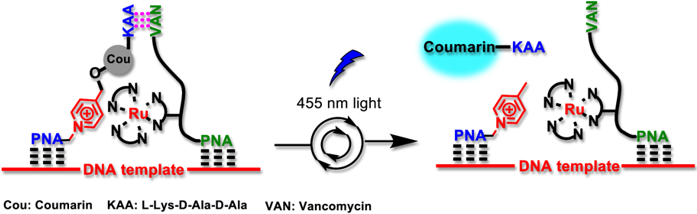
Overcoming product inhibition and facilitating template turn-over using a 3-way junction
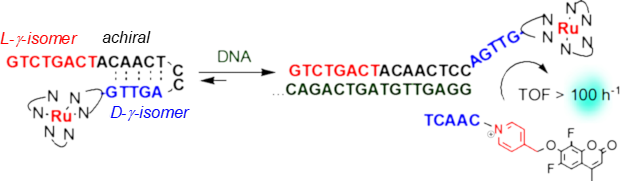
Templated-formation of ruthenium-based photocatalyst

Coupling bioluminescence to templated photocatalytic reactions
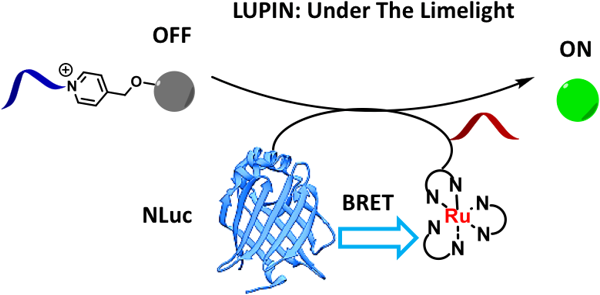
CHEMICAL BIOLOGY PROBES, BIOACTIVE NATURAL PRODUCTS AND MEDICINAL CHEMISTRY
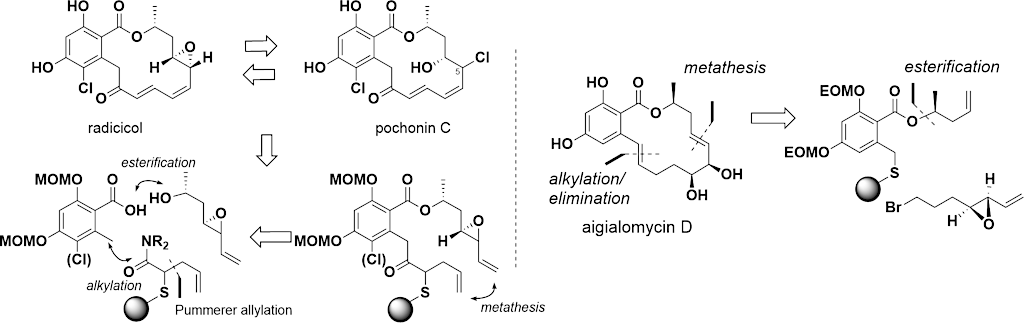
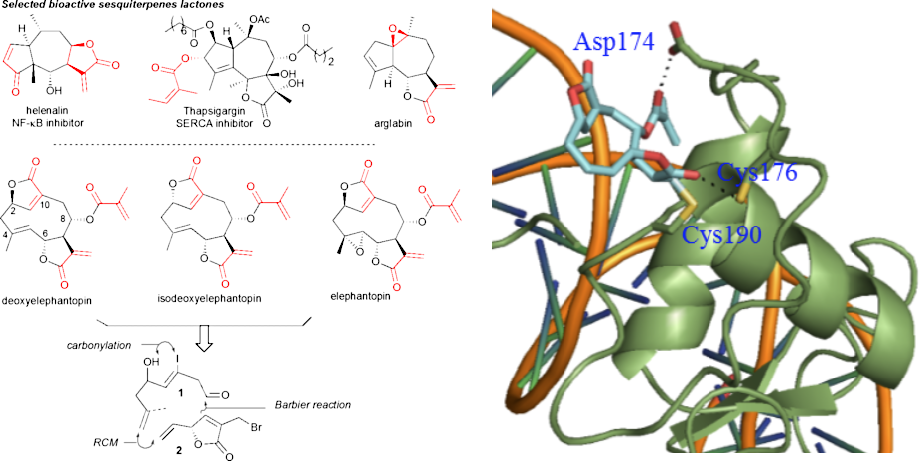
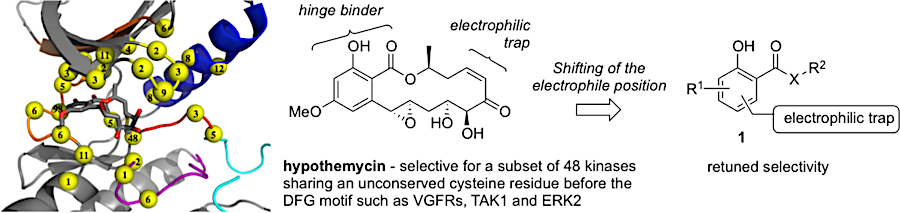
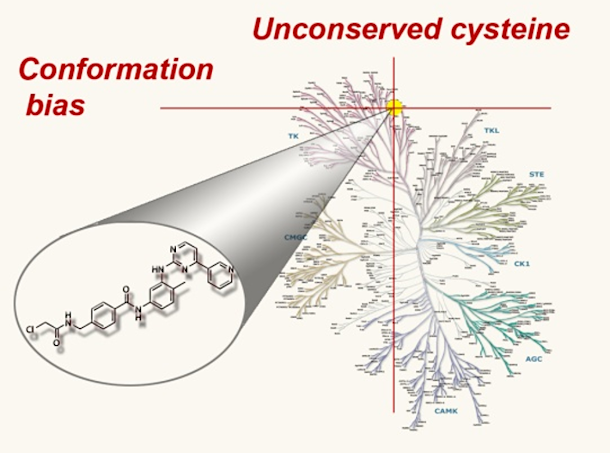
Bioactive natural products offer a privileged starting point for the design of chemical biology probes and to develop synthesis method that enable faster or easier access to important structural motifs. This research line has led to the discovery of several important inhibitors and tool compounds for chemical biology: a potent HSP90 inhibitor that highlighted a previously unrecognized binding pocket within HSP90 and served to uncover the role of the HSP90α in spermatocyte development; resorcylic acid lactones that covalent inhibitor that targets the angiogenic kinases (VEGFRs); analogs were used to dissect the regulatory fate of a signaling kinase TAK1; the first covalent inhibitor of KIT kinase designed based on a mapping of unconserved cysteines in the kinome. Our interest in covalent inhibitors also led us to investigate sesquiterpene lactones which frequently harbor mildly reactive functionalities that can engage with a nucleophilic residue (most often cysteine) in the target. Synthesis of bioactive probe allowed us to uncover a potent covalent inhibitor of PPAR-gamma.
Synthesis of deoxyelephantopin probes to uncover its mode of action
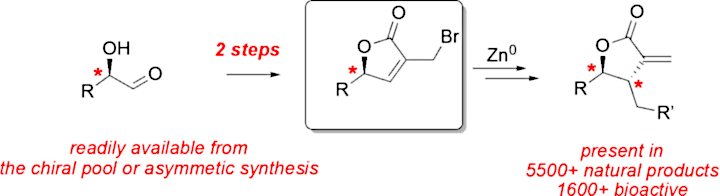
Fast and simple strategy to access chiral butyrolactone
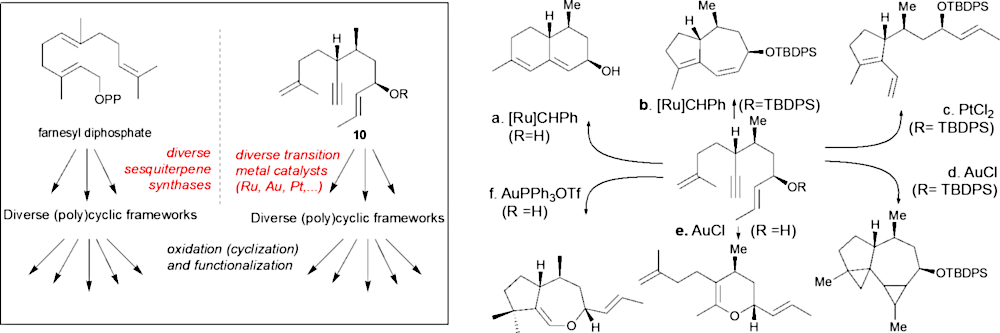
Diversity-oriented cyclization cascade inspired by sesquiterpene biosynthesis
Retuning the kinase selectivity of resorcylic acid lactones
Designed covalent kinase inhibitor
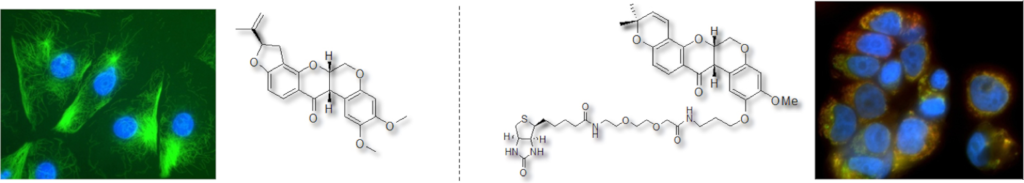
Synthesis of deguelin and rotenone probes to investigate their mode of action
Synthetic strategy for rapid access to resorcylic acid lactones