Research groups
Current Selection : 25 Research Groups
Analytical Chemistry, Electrochemistry, Electrochemical Sensors, Optical Detection Principles, Ion Optodes, Exhaustive Sensors, Photoelectric Conversion, Light Activated Extraction, Nanosphere Reagents, Materials Characterization, Polymers and Polymer Modifications, Environmental Analysis, Biomedical Analysis.
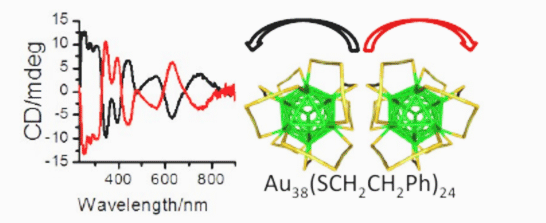
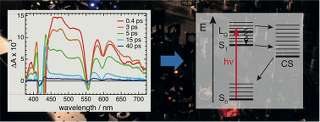
Chemistry, spectroscopy and applications of surfaces, interfaces and chiral nanomaterials: Development and application of spectroscopic techniques to probe solid-liquid interfaces, preparation and applications of chiral nanomaterials, enantiodifferentiation, vibrational optical activity, self-assembly of plasmonic nanomaterials.
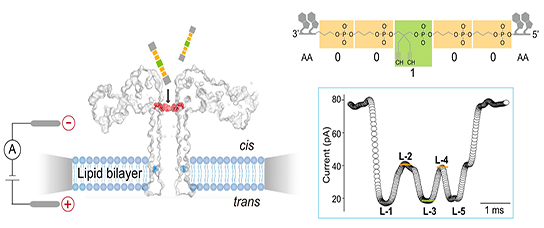
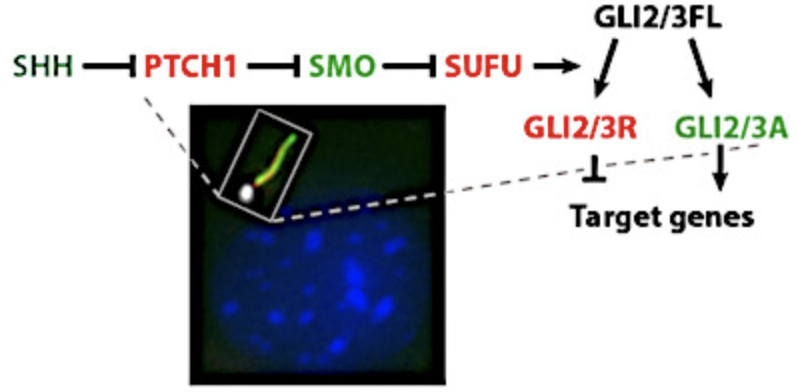
We want to understand in physical and molecular terms how cells talk to each other during development. This means our research is highly interdisciplinary: physics, cell biology, molecular biology, biochemistry, genetics... Indeed some of us in the lab are biologists, other physicists, chemists, engineers.
We are interested in the signaling events that control tissue growth: how is the shape and final size of a tissue achieved during embryogenesis?
We focus on two types of proliferation modes: growth control by morphogen gradients and asymmetric cell division in stem cells. We do this using two model systems: Drosophila and Zebrafish.
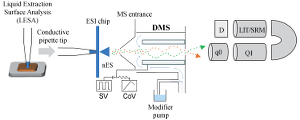
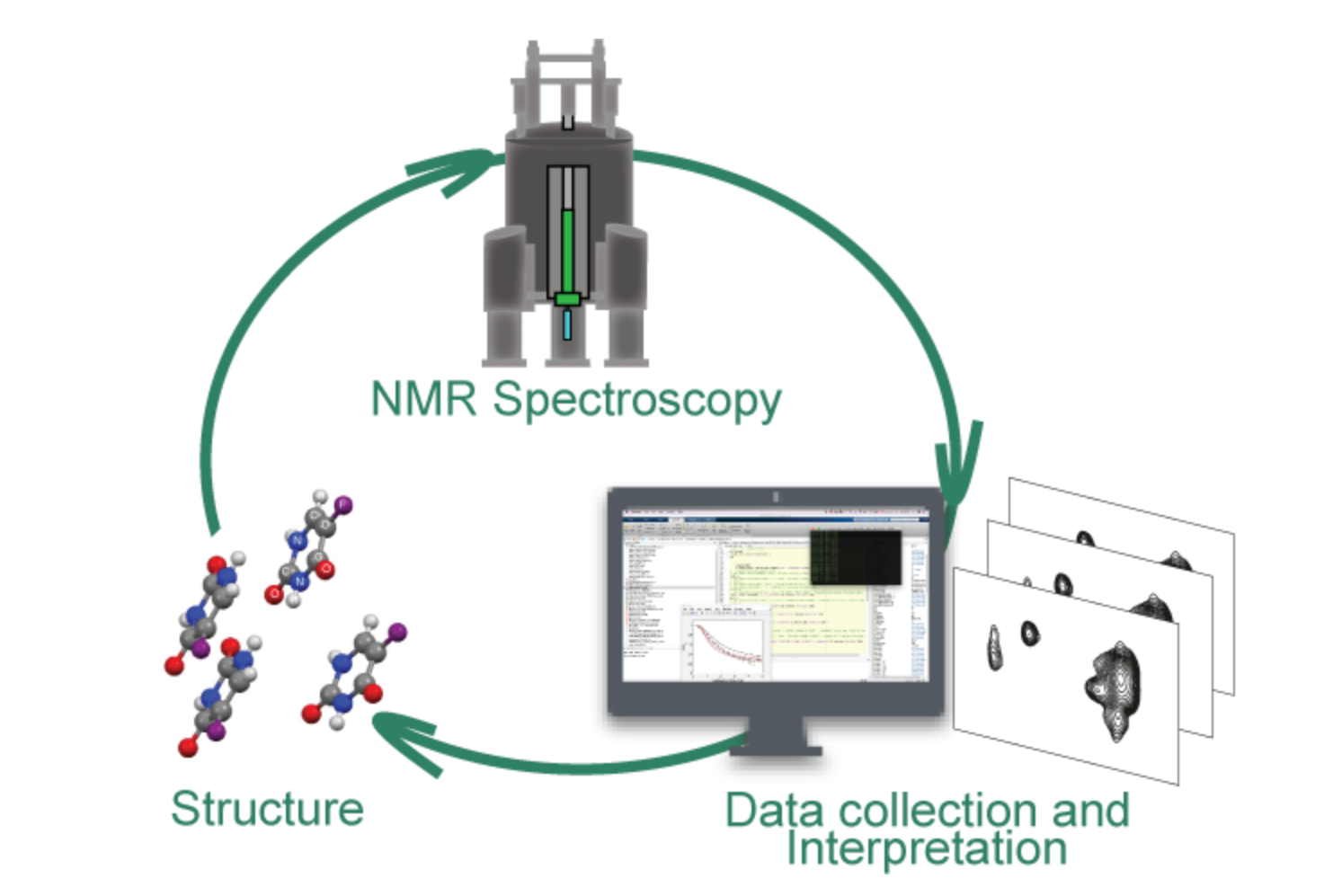
Analytical chemistry and mass spectrometry of low molecular weight compounds (pharmaceuticals) and macromolcules (peptides, proteins). Mass spectrometry imaging, Ionization, High resolution MS and data independent acquisition, ion mobility mass spectrometry, coupling MS with separation sciences, data analysis, MS libraries, bioanalysis, metabolism, metabolomics, lipidomics, proteomics, ultra-fast quantitative analysis.
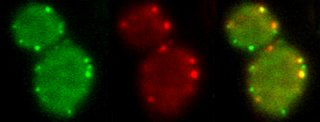
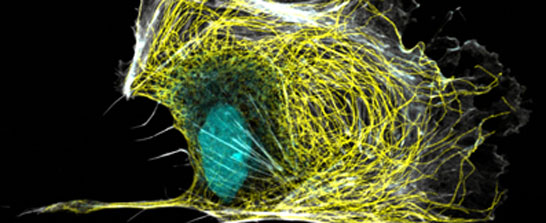
We study the molecular mechanisms of membrane traffic, especially clathrin-mediated endocytosis. We aim to understand the assembly, function and regulation of the complex molecular machineries that drive the formation of endocytic vesicles. Our main experimental organism is budding yeast. We use a combination of quantitative live-cell imaging, electron microscopy, genetics, biochemistry and structural biology.
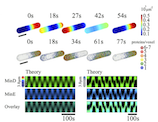
We study the formation of spatial and temporal structures in individual biological cells and cells assemblies. The focus of our work is on theoretical descriptions of cytoskeletal dynamics. The cytoskeleton is a network of filamentous proteins, which is kept permanently out of thermodynamic equilibrium. It enables cells to divide, determines their shape and plays an important role in cell locomotion. In our descriptions, we rely heavily on concepts from non-linear dynamics and from non-equilibrium statistical mechanics.


My research group focuses on understanding how mechanics of lipid membranes can influence the life of cells. The enveloppe of living cells is made of lipid bilayers which impermeability and deformability ensure changes in cell shape while keeping its specific content. We are interested in understanding how membrane mechanical properties can constraint several cell processes at the molecular, cellular and multi-cellular scales. In particular, we focused on how membrane tension and rigidity influence intracellular membrane traffic (in particular endocytosis and Golgi trafficking) and cell division (in particular cytokinesis). We recently started to be interested in multi-cellular systems, in particular epithelia, where membrane tension is proposed to play a role in cell reorganization during organ morphogenesis.
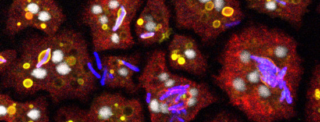
The major aim of my group is to understand the integration of signalling, cytoskeleton and membrane trafficking in phagocytosis and its relevance to host-pathogen interactions. To this end, we use the social amoeba Dictyostelium as a model organism as it is a is genetically and biochemically tractable professional phagocyte very similar to phagocytes of the innate immune system in morphology and behaviour.